auspice export
In [1]:
import baltic as bt
import requests
from io import StringIO as sio
from datetime import datetime as dt
import json
def convertToJSON(node,most_recent_tip,index=None,annotate_nodes=None):
"""
Traverse baltic tree and create a json object
"""
if index==None: index=0
json_node={'name': None,
'node_attrs': {'num_date': {'value': node.absoluteTime}},
'branch_attrs': {}
} ## basic json branch with num_date (absolute time) assigned
if 'height_95%_HPD' in node.traits: ## height 95% HPD available, compute from most recent tip date
lower,upper=node.traits['height_95%_HPD'] ## get height 95% HPD (counted here as years backwards from most recent tip)
time_range = [most_recent_tip-upper, most_recent_tip-lower] ## format range
json_node['node_attrs']['num_date']['confidence']=time_range ## assign 95% HPD for height to node
if annotate_nodes: ## attempting to fetch more traits
for attr in annotate_nodes: ## iterate over attributes
if attr in node.traits: ## attribute available for baltic node
json_node['node_attrs'][attr]=annotate_nodes[attr](node) ## annotate
if node.is_node(): ## node
json_node['children']=[] ## has children
json_node['name']='NODE_%07d'%(index) ## different name
for child in node.children: ## iterate over children
if child.is_node(): index+=1 ## increment index if child is node too
index,json_child=convertToJSON(child,most_recent_tip,index=index,annotate_nodes=annotate_nodes) ## get the json-formatted child
json_node['children'].append(json_child) ## attach resulting json-formatted children to current json node
else:
json_node['name']=node.name ## leaf, name is simple
return index,json_node
def toNextstrainJSON(tree,output,meta,annotate_nodes=None):
"""
Convert a baltic tree object to JSON, output to file
"""
out_file=open(output,'w')
_,json_tree=convertToJSON(tree.root,tree.mostRecent,annotate_nodes=annotate_nodes)
meta['tree']=json_tree
output_json=meta
json.dump(output_json,out_file,indent=1) ## write to file
out_file.close()
def generic_continuous_interval(node,trait,which='_95%_HPD'):
"""
Given a continuous trait and a baltic branch object convert from beast to auspice format.
"""
return_dict={}
return_dict['value']=node.traits[trait]
if '%s%s'%(trait,which) in node.traits:
return_dict['confidence']=node.traits['%s%s'%(trait,which)]
return return_dict
def generic_categorical_import(node,trait):
"""
Given a discrete trait and a baltic branch object convert from beast to auspice format.
"""
return_dict={}
return_dict['value']=node.traits[trait]
if '%s.set'%(trait) in node.traits:
return_dict['confidence']={t: p for t,p in zip(node.traits['%s.set'%(trait)],node.traits['%s.set.prob'%(trait)])}
return return_dict
In [2]:
address='https://github.com/sdwfrost/mers-treedater/raw/master/MERS_274_sCoal.combinedTyped.mcc.tree' ## where the tree we'll use lives
fetch_tree = requests.get(address) ## fetch tree
treeFile=sio(fetch_tree.text) ## stream from repo copy
ll=bt.loadNexus(treeFile) ## treeFile here can alternatively be a path to a local file
ll.treeStats()
### the basic auspice template listing:
## colour-by's
## panels
## display defaults
## filters
meta={'version': 'v2',
'meta': {'updated': '%s'%(dt.strftime(dt.now(),'%Y-%m-%d')),
'colorings': [{'key': 'type', 'title': 'Host', 'type': 'categorical'},
{'key': 'posterior', 'title': 'Posterior', 'type': 'continuous'},
{'key': 'num_date', 'title': 'Date', 'type': 'continuous'}],
'panels': ['tree'],
'display_defaults': {'color_by': 'type',
'distance_measure': 'num_date'},
'filters': ['type']
}
}
### dict that will annotate json by accessing traits of a baltic tree
annotate_nodes={'type': lambda k: generic_categorical_import(k,'type'),
'posterior': lambda k: {'value': k.traits['posterior']}
}
out='MERS_auspice.json'
toNextstrainJSON(ll,out,meta,annotate_nodes)
Tree height: 5.651292 Tree length: 79.196209 strictly bifurcating tree annotations present Numbers of objects in tree: 547 (273 nodes and 274 leaves)
In [3]:
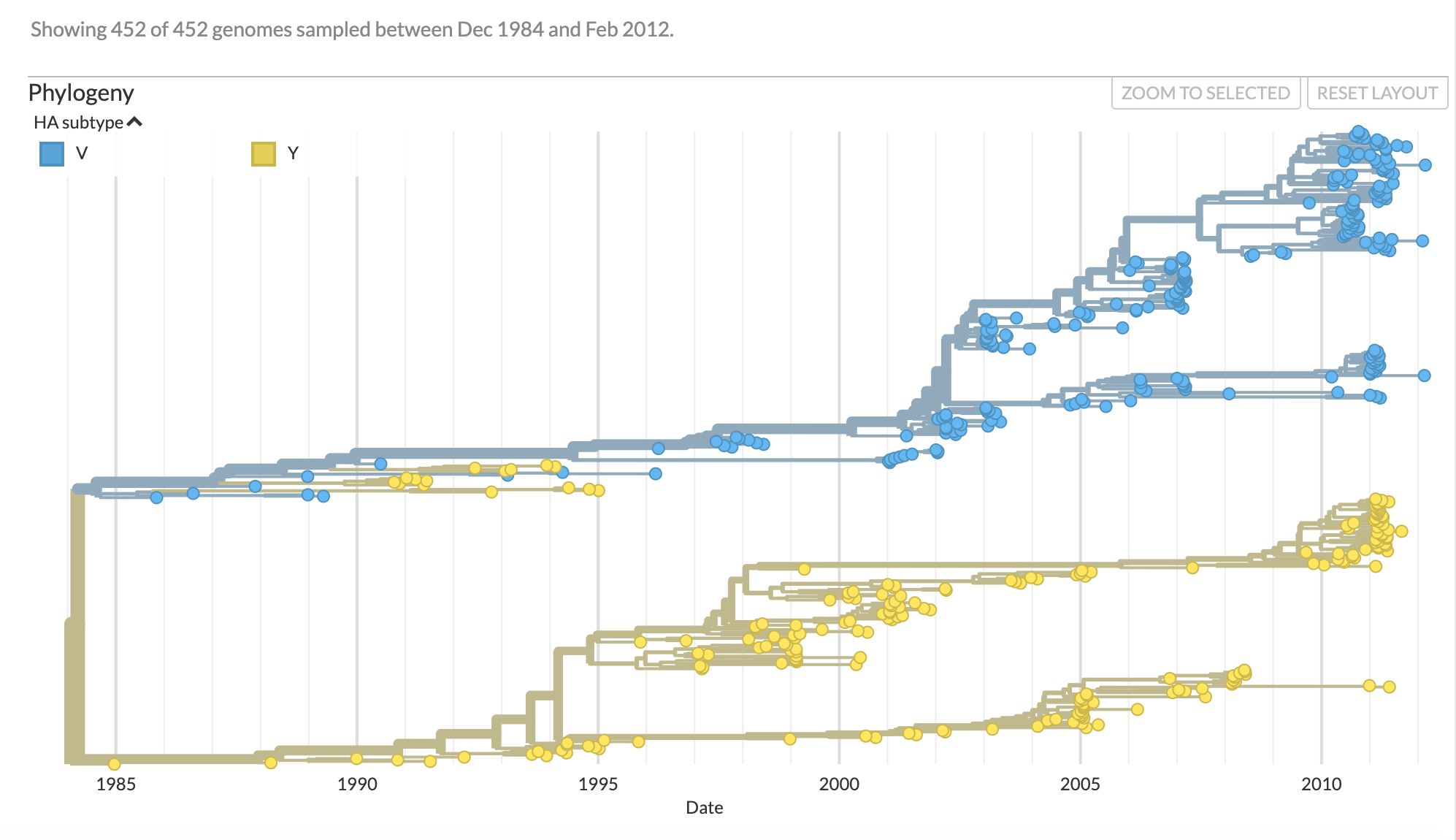
address='https://raw.githubusercontent.com/evogytis/fluB/master/data/mcc%20trees/InfB_PB1t_ALLs1.mcc.tre' ## address of example tree
fetch_tree = requests.get(address) ## fetch tree
treeFile=sio(fetch_tree.text) ## stream from repo copy
ll=bt.loadNexus(treeFile,tip_regex='_([0-9\-]+)$') ## treeFile here is a path to a local file, with collection dates encoded in tip names (numbers and hyphens at the end, found by regex)
# ll.setAbsoluteTime(bt.decimalDate('2012-02-26')) ## if tip names don't contain an encoded collection date just give the most recent date to set absolute times in tree
ll.treeStats() ## report stats about tree
meta={'version': 'v2',
'meta': {'updated': '%s'%(dt.strftime(dt.now(),'%Y-%m-%d')),
'colorings': [{'key': 'PB2', 'title': 'PB2 subtype', 'type': 'categorical'},
{'key': 'PA', 'title': 'PA subtype', 'type': 'categorical'},
{'key': 'NP', 'title': 'NP subtype', 'type': 'categorical'},
{'key': 'HA', 'title': 'HA subtype', 'type': 'categorical'},
{'key': 'NS', 'title': 'NS subtype', 'type': 'categorical'},
{'key': 'MP', 'title': 'MP subtype', 'type': 'categorical'},
{'key': 'posterior', 'title': 'Posterior', 'type': 'continuous'},
{'key': 'num_date', 'title': 'Date', 'type': 'continuous'},
{'key': 'S', 'title': 'Synonymous mutations', 'type': 'continuous'},
{'key': 'N', 'title': 'Non-synonymous mutations', 'type': 'continuous'}],
'panels': ['tree'],
'display_defaults': {'color_by': 'HA',
'distance_measure': 'num_date'},
'filters': ['PB2','PA','NP','HA','NS','MP']
}
}
### dict that will annotate json by accessing traits of a baltic tree
annotate_nodes={'PB2': lambda k: generic_categorical_import(k,'PB2'),
'HA': lambda k: generic_categorical_import(k,'HA'),
'NP': lambda k: generic_categorical_import(k,'NP'),
'PA': lambda k: generic_categorical_import(k,'PA'),
'NS': lambda k: generic_categorical_import(k,'NS'),
'MP': lambda k: generic_categorical_import(k,'MP'),
'posterior': lambda k: {'value': k.traits['posterior']},
'N': lambda k: generic_continuous_interval(k,'S',which='_95%_HPD'),
'S': lambda k: generic_continuous_interval(k,'N',which='_95%_HPD'),
'div': lambda k: k.traits['div']
} ## keys access baltic traits
for k in ll.Objects: ## create new trait for baltic tree that will compute (mean) number of mutations for branch (substitution length)
cur_node=k
k.traits['div']=0.0 ## assume zero branch length
while cur_node: ## while not at root
k.traits['div']+=cur_node.traits['N']+cur_node.traits['S'] if ('N' in cur_node.traits or 'S' in cur_node.traits) else 0.0 ## continue adding mutation length
cur_node=cur_node.parent ## go to parent
out='fluB_auspice.json'
toNextstrainJSON(ll,out,meta,annotate_nodes)
Tree height: 28.071359 Tree length: 481.049928 strictly bifurcating tree annotations present Numbers of objects in tree: 903 (451 nodes and 452 leaves)
In [ ]: