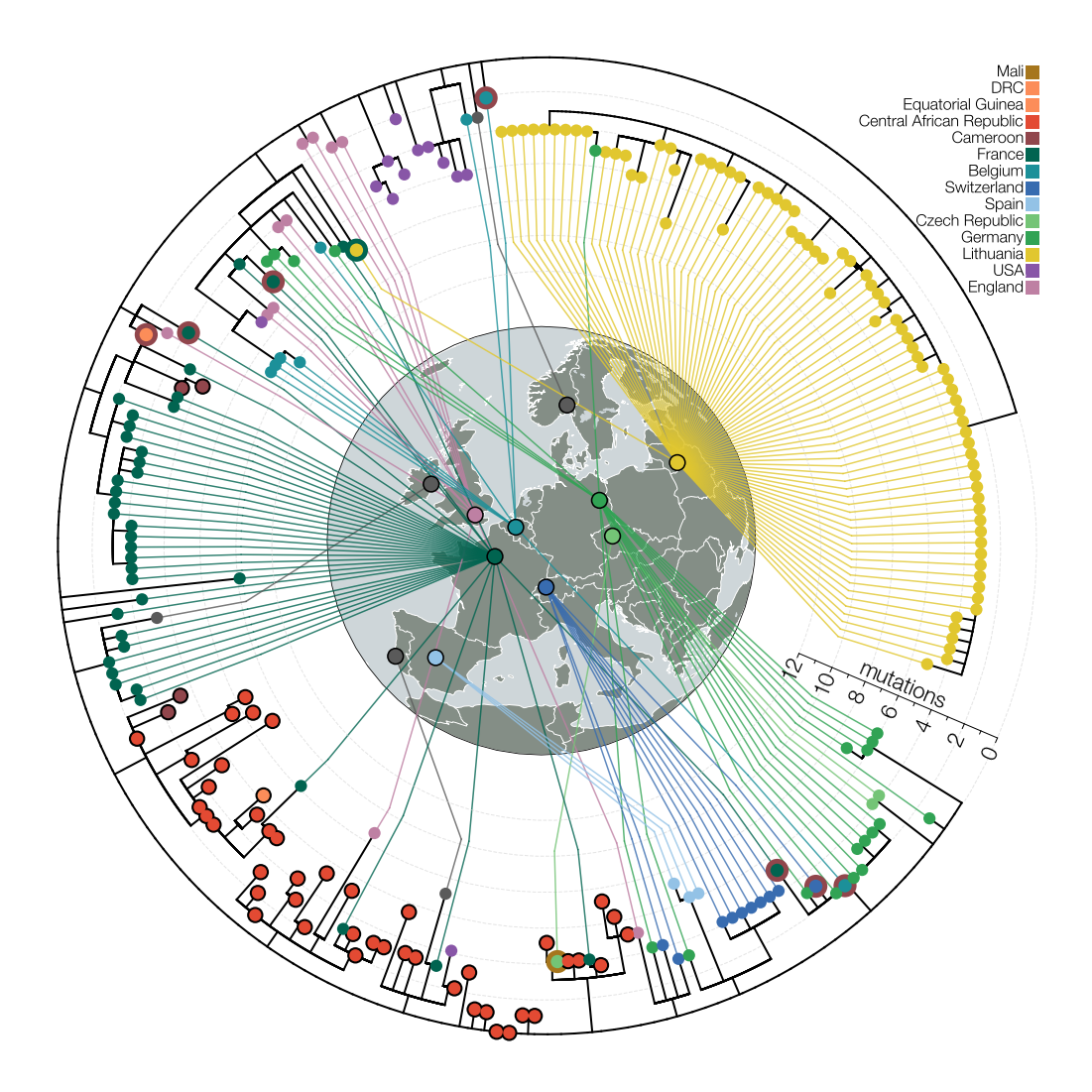
circular tree surrounding map
In [1]:
%matplotlib inline
import matplotlib as mpl
from matplotlib import pyplot as plt
import matplotlib.patheffects as path_effects
from matplotlib.collections import LineCollection
from matplotlib.patches import Rectangle,ConnectionPatch
from matplotlib.gridspec import GridSpec
typeface='Helvetica Neue'
mpl.rcParams['font.weight']=300
mpl.rcParams['axes.labelweight']=300
mpl.rcParams['font.family']=typeface
mpl.rcParams['font.size']=22
mpl.rcParams['pdf.fonttype']=42
import os,glob,requests
from io import StringIO as sio
import numpy as np
import baltic as bt
import cartopy
import cartopy.crs as ccrs
### this example uses data from a study on B.1.620, a SARS-CoV-2 lineage with multiple VOC-like mutations and deletions discovered in Lithuania
# the story of lineage B.1.620 can be found here: https://evogytis.github.io/posts/2021/10/B1620/
# you can find the published version here: https://www.nature.com/articles/s41467-021-26055-8
address='https://raw.githubusercontent.com/phylo-baltic/baltic-gallery/gh-pages/assets/data/B_1_620-baltic.newick'
fetch_tree = requests.get(address) ## fetch tree
treeFile=sio(fetch_tree.text) ## stream from repo copy
ll=bt.loadNewick(treeFile) ## treeFile here can alternatively be a path to a local file
ll.treeStats() ## report stats
alnL=29903 ## alignment length for rescaling tree into raw mutations
ll=ll.collapseBranches(lambda k: k.length==0.0) ## collapse arbitrarily resolved polytomies into actual polytomies
for k in ll.Objects:
k.length=k.length*alnL ## rescale branches to mutations
ca=ll.getExternal(lambda k: k.name=='hCoV-19/Central_African_Republic/234/2021||2021-03-31')[0].parent ## get common ancestor of all B.1.620
ll=ll.subtree(ca) ## prune to subtree
ll.root.length=0.1 ## a tiny bit of branch length on the root
ll.traverse_tree() ## make sure heights are set correctly
ll.treeStats() ## report new stats on collapsed and pruned tree
coordinates={'Cameroon': (3.844119,11.501346),
'England': (51.509865,-0.118092),
'Norway': (59.91273, 10.74609),
'DRC': (-4.322447,15.307045),
'Ireland': (53.33306, -6.24889),
'Belgium': (50.850346,4.351721),
'Central_African_Republic': (4.36122,18.55496),
'Czech_Republic': (50.073658,14.418540),
'Switzerland': (46.947456,7.451123),
'USA': (38.889248,-77.050636),
'Lithuania': (54.68916, 25.2798),
'Germany': (52.531677,13.381777),
'Equatorial_Guinea': (3.75578,8.78166),
'Portugal': (38.736946,-9.142685),
'France': (48.864716,2.349014),
'Spain': (40.4165, -3.70256)} ## coordinates for places in the tree
travel={'hCoV-19/Belgium/rega-5050/2021|EPI_ISL_1382294|2021-03-14': 'Cameroon',
'hCoV-19/France/HDF-P326-21134M0692/2021|EPI_ISL_1671822|2021-03-31': 'Cameroon',
'hCoV-19/Equatorial_Guinea/91946/2021|EPI_ISL_1673323|2021-02-06': 'Cameroon',
'hCoV-19/Czech_Republic/183304/2021|EPI_ISL_1675656|2021-03-25': 'Mali',
'hCoV-19/France/ARA-210013001901/2021|EPI_ISL_1406653|2021-02-26': 'Cameroon',
'hCoV-19/Switzerland/GE-33576177/2021|EPI_ISL_1369646|2021-03-16': 'Cameroon',
'hCoV-19/Belgium/ULG-12917/2021|EPI_ISL_1241728|2021-03-01': 'Cameroon',
'hCoV-19/Lithuania/MR-LUHS-5-8/2021|EPI_ISL_1576950|2021-03-26': 'France',
'hCoV-19/France/PDL-IPP07069/2021|EPI_ISL_1495980|2021-03-18': 'Cameroon'} ## information about tips that are from travel cases
colours={'England': '#BE80A3',
'USA': '#8856a7',
'Lithuania': '#E1C72F',
'Germany': '#31a354',
'Czech_Republic': '#74c476',
'Spain': '#5A5A5A',
'Switzerland': '#386cb0',
'Belgium': '#1c9099',
'France': '#016450',
'Cameroon': '#91464C',
'Central_African_Republic': '#e34a33',
'Equatorial_Guinea': '#fc8d59',
'DRC': '#fc8d59',
'Mali': '#a6761d',
'Spain': '#93C2E6',
'Norway': '#5A5A5A',
'Ireland': '#5A5A5A',
'Portugal': '#5A5A5A'} ## colours for all the locations
Tree height: 0.001275 Tree length: 0.029111 strictly bifurcating tree Numbers of objects in tree: 595 (297 nodes and 298 leaves) Tree height: 10.177311 Tree length: 314.231015 Numbers of objects in tree: 311 (74 nodes and 237 leaves)
In [2]:
def polar_transform(ax,tree,circStart=0.0,circFrac=1.0,inwardSpace=0.0,precision=20):
"""
Given axes and a baltic tree object plot a circular tree.
"""
x_attr=lambda k:k.x
y_attr=lambda k:k.y
colour='k'
width=2
if inwardSpace<0: inwardSpace-=tree.treeHeight ## if inward space is negative then tree is pointing inwards
branches=[] ## will hold branch coordinates
cs=[] ## will hold colours
linewidths=[] ## will hold branch widths
circ_s=circStart*np.pi*2 ## where circle starts
circ=circFrac*np.pi*2 ## how long the circle is (2pi=full)
allXs=list(map(x_attr,tree.Objects)) ## get all branch heights
allXs.append(max(allXs)*1.1) ## add a guaranteed maximum height that's 10% bigger
normaliseHeight=lambda value: (value-min(allXs))/(max(allXs)-min(allXs)) ## create normalisation function for height
linspace=lambda start,stop,n: list(start+((stop-start)/(n-1))*i for i in range(n)) if n>1 else stop ## hacky linearly spaced list of floats function
done=[] ## remember which coordinates were done on the map
### scale bar
scale_bar_length=12
y0=circ_s-0.1 ## where scale bar will begin - where tree starts, but pulled back a bit
x0=normaliseHeight(0+inwardSpace) ## scale bar starts at 0 (+offset)
x1=normaliseHeight(scale_bar_length+inwardSpace) ## scale bar finishes at 12 (+offset)
### scale bar main line
ax.plot([np.sin(y0)*x0,np.sin(y0)*x1],[np.cos(y0)*x0,np.cos(y0)*x1],color='k',lw=1)
#### scale bar ticks
for i in range(0,scale_bar_length+1):
w=0.01+i*0.0005 ## tick size increases towards centre
y_line=np.linspace(y0,y0+w,10) if i%2==0 else np.linspace(y0,y0+w/1.5,10) ## 10 segments to curve the ticks slightly, size alternates between odd and even
x=normaliseHeight(i+inwardSpace) ## tick position
ax.plot(np.sin(y_line)*x,np.cos(y_line)*x,color='k',lw=1) ## plot tick
if (i)%2==0: ## every even tick
ax.plot(np.sin(np.linspace(-np.pi,np.pi,100))*x,np.cos(np.linspace(-np.pi,np.pi,100))*x,color='lightgrey',ls='--',alpha=0.6,lw=1,zorder=0) ## plot faint full circle for labelled ticks
ax.text(np.sin(y_line[-1]+w*2.4)*x,np.cos(y_line[-1]+w*2.4)*x,'%d'%(i),ha='center',va='center',rotation=-np.rad2deg(y_line[-1])) ## add tick label
#### scale bar label
ax.text(np.sin(y0-0.03)*np.mean([x0,x1]),np.cos(y0-0.03)*np.mean([x0,x1]),'mutations',ha='center',va='center',rotation=-np.rad2deg(y0)-90)
for k in tree.Objects: ## iterate over branches
x=normaliseHeight(x_attr(k)+inwardSpace) ## get branch x position
xp=normaliseHeight(x_attr(k.parent)+inwardSpace) if k.parent.parent else x ## get parent x position
y=y_attr(k) ## get y position
try: ## try getting colour
cs.append(colour(k)) if callable(colour) else cs.append(colour)
except KeyError: ## grey if failed
cs.append((0.7,0.7,0.7))
linewidths.append(width(k)) if callable (width) else linewidths.append(width) ## add branch width
y=circ_s+circ*y/tree.ySpan ## convert y coordinate
X=np.sin(y) ## polar transform
Y=np.cos(y) ## polar transform
branches.append(((X*xp,Y*xp),(X*x,Y*x))) ## add branch segment
if k.is_node(): ## internal node
yl,yr=y_attr(k.children[0]),y_attr(k.children[-1]) ## get leftmost and rightmost children's y coordinates
yl=circ_s+circ*yl/tree.ySpan ## transform y into a fraction of total y for left child
yr=circ_s+circ*yr/tree.ySpan ## same for right child
ybar=linspace(yl,yr,precision) ## what used to be vertical node bar is now a curved line
xs=[yx*x for yx in np.sin(ybar)] ## convert to polar coordinates
ys=[yy*x for yy in np.cos(ybar)] ## convert to polar coordinates
branches+=tuple(zip(zip(xs,ys),zip(xs[1:],ys[1:]))) ## add curved segment
linewidths+=[linewidths[-1] for q in zip(ys,ys[1:])] ## repeat linewidths
cs+=[cs[-1] for q in zip(ys,ys[1:])] ## repeat colours
else: ## external node/tip
strain=k.name#.split('|')[0] ## get name
country=strain.split('/')[1]
if country in coordinates: ## coordinate available for tip
lat,lon=coordinates[country] ## fish out coordinates for tip
s=150 ## size of marker
country=strain.split('/')[1] ## get country
fc=colours[country] ## get colour for tip
ec='k' ## get edge colour for tip
scale=1.8 ## set edge line size
if strain in travel: ## tip is from a traveller
s*=1.2 ## increase marker size
scale=3 ## increase edge line size
ec=colours[travel[strain]] ## get new edge colour
ax.scatter(X*x,Y*x,s=s,facecolor=fc,edgecolor='none',zorder=10000) ## plot main marker
if strain in travel or country in ['Central_African_Republic','Cameroon','Equatorial_Guinea', 'DRC']: ## only mark the outside of the tip if it's a traveller or an African sequence
ax.scatter(X*x,Y*x,s=s*scale,facecolor=ec,edgecolor='none',zorder=9999) ## tip circle
if country in ['Central_African_Republic','Equatorial_Guinea', 'DRC','Cameroon','USA']: ## don't plot locations not in Europe
pass
else: ## tip in Europe, plot location on a map, connect to tree with a line
# print(locs[strain])
latT,lonT=ortho.transform_point(lon, lat, ccrs.PlateCarree()) ## get location of tip in current projection coordinates
al=0.8 ## connection lines will be slightly transparent
lw=1.5 ## connection lines will be slightly thicker than regular
ls='-' ## connection lines entire
treeEnd=normaliseHeight(ll.treeHeight*1.01+inwardSpace) ## coordinate that sticks out a bit past the tree's highest point
ax.plot([X*treeEnd,X*x],[Y*treeEnd,Y*x],color=fc,ls=ls,lw=lw,alpha=al) ## draw line departing each tip and going off to some high point
con = ConnectionPatch(xyA=(X*treeEnd,Y*treeEnd), ## connect end of the line that departed a tip
coordsA=ax.transData, ## coordinate provided is in dat aspace
axesA=ax, ## tree part of the plot
xyB=(latT,lonT), ## connect to map
coordsB=ax2.transData, ## in map coordinates
axesB=ax2, ## map part of the plot
color=fc, ls=ls,lw=lw,zorder=3,alpha=al) ## colour, line style, linewidth, order, transparency
ax2.add_patch(con) ## add line to plot
s=200 ## square marker size
if (lon,lat) not in done: ## haven't done map coordinate before
ax2.scatter(lon,lat,s=s,facecolor=fc,edgecolor='none', ## plot circle on map
zorder=10000,transform=ccrs.PlateCarree(),clip_on=False)
ax2.scatter(lon,lat,s=1.5*s,facecolor='k',edgecolor='none', ## plot outline circles on map
zorder=9999,transform=ccrs.PlateCarree(),clip_on=False)
done.append((lon,lat)) ## remember coordinate was done
line_segments = LineCollection(branches,lw=linewidths,ls='-',color=cs,capstyle='projecting',zorder=2) ## create line segments that will be tree branches
ax.add_collection(line_segments) ## add collection to axes
line_segments = LineCollection(branches,lw=[lw*3 for lw in linewidths],ls='-',color=['w' for c in cs],capstyle='projecting',zorder=1) ## create line segments that will be a white outline to tree branches
ax.add_collection(line_segments) ## add collection to axes
In [3]:
fig,ax = plt.subplots(figsize=(20,20),facecolor='w')
gs = GridSpec(1,1,hspace=0.01,wspace=0.0)
ax=plt.subplot(gs[0],zorder=1000,facecolor='none')
############
ortho=ccrs.NearsidePerspective(central_longitude=7,central_latitude=50,satellite_height=800000) ## projection
w=0.3
ax2=fig.add_axes([(1-w)/2, (1-w)/2, w, w],facecolor='none',projection=ortho,zorder=0) ## add sub-axes
[ax2.spines[loc].set_visible(False) for loc in ax.spines] ## no spines for the plot
p1 = ax.get_position()
p2 = ax2.get_position()
scale=2
w=0.3
ax2.set_position([(1-w)/2+0.009, (1-w)/2+0.006, w, w]) ## adjust position of map subplot, it's not perfectly centered
## map scale and colour
scale='50m'
water='#CED6D9'
land='#848E86'
# water='w'
# land='#C2C4BD'
## add water bodies, continents and country borders
ax2.add_feature(cartopy.feature.LAKES.with_scale(scale),facecolor=water)
ax2.add_feature(cartopy.feature.OCEAN.with_scale(scale),facecolor=water,edgecolor=water)
ax2.add_feature(cartopy.feature.LAND.with_scale(scale),facecolor=land,edgecolor='w')
ax2.add_feature(cartopy.feature.BORDERS.with_scale(scale),edgecolor='w',lw=1,zorder=1)
country_order=['Lithuania','Belgium','Norway','USA','England','DRC','Cameroon','France','Central_African_Republic','Equatorial_Guinea','Ireland','Switzerland','Spain','Portugal','Czech_Republic','Germany'] ## indices of each country are used as (mean) weights to sort children of nodes in the tree, adjust to get desired order of nodes in the tree
ll.root.children=sorted(ll.root.children,key=lambda w: np.mean([country_order.index(lf.split('|')[0].split('/')[1]) for lf in w.leaves]) if w.is_node() else country_order.index(w.name.split('|')[0].split('/')[1])) ## sort children of root based on mean index of each child
ll.drawTree() ## recompute coordinates of branches after sorting
polar_transform(ax,ll,circStart=0.83,circFrac=0.97,inwardSpace=-17,precision=200) ## plot tree in polar coordinates
## remove ticks and tick labels from everything
ax.set_xticks([])
ax.set_xticklabels([])
ax.set_yticks([])
ax.set_yticklabels([])
ax2.set_xticks([])
ax2.set_xticklabels([])
ax2.set_yticks([])
ax2.set_yticklabels([])
[ax.spines[loc].set_visible(False) for loc in ax.spines] ## no spines for tree plot
ax.set_aspect(1)
## legend
h=100000
w=100000
for c,country in enumerate(list(colours.keys())[::-1]): ## iterate over specific countries
if colours[country]!='#5A5A5A':
x=3500000 ## where legend patch starts horizontally
y=3700000-c*h*1.2 ## where legend patch starts vertically
fc=colours[country] ## get colour
effects=[path_effects.Stroke(linewidth=4, foreground='white'),
path_effects.Stroke(linewidth=0, foreground='k')] ## black text, white outline
ax2.add_patch(Rectangle((x,y),w,h,facecolor=fc,edgecolor='none',clip_on=False)) ## add coloured legend patch
ax2.text(x-w*0.1,y+h/2,country.replace('_',' '),color='k',size=16,va='center',ha='right',path_effects=effects,zorder=1000000) ## add text label
plt.show()
In [ ]: